-
Notifications
You must be signed in to change notification settings - Fork 20
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Workflow - Scientist #16
Comments
|
A minimal git repository that would work as "executable paper" for the "Great icecream preference study of 2016": The We can provide a the |
|
ping @ctb (who should start watching this repo for all the notifications all the time) |
|
What you would see as the publication is the rendered version of |
|
The entry point would always be We should provide a base docker image that contains
|
|
Yes, good stuff! My concern is that if this is the only allowed structure and workflow (as opposed to merely a strongly recommended one) we will automatically lose most potential early adopters - essentially, anyone who is already doing their own thing in this area. With the specfile idea we could allow a much broader range of repo structure/workflow (and provide a Web site to build the spec by inspecting a repo), while using the above as a specific structure & workflow for demo purposes. |
|
A strong -1 on it being a shell script - something declarative offers many more opportunities for simplicity and introspection and composition. If procedural (like a Dockerfile or a shell script) then we need to run it to find out what it does. With a YAML spec, it could specify what resources need to be present, along with inputs and outputs, and then everything (travis.yml) could be produced from that, no? |
|
If you are doing your own thing, and don't want to make a We could also inject the required stuff via docker-compose. This might remove the need for a shared base image. |
|
On Tue, Feb 23, 2016 at 06:30:28AM -0800, Tim Head wrote:
that can be part of the spec, no? We can require it be md or md-convertible,
Yep! |
|
Specifying resources is a pro for having Con for docker-compose, we need to know which version of the required-stuff image to inject. Could be noted in I am open for supporting more formats for people to write their paper in. I would insist though that the format they use has a way of mixing code with prose (like a notebook). For markdown and ipynb I know how to do that. Do you know (sane) ways of doing this in LaTeX? After reflecting on this over ☕ I am 👍 on a |
|
On Tue, Feb 23, 2016 at 06:42:21AM -0800, Tim Head wrote:
Agreed on world of pain! And agree we should allow arbitrary config (perhaps
coo'. |
|
(note to future: in the above comment there is a sentence from titus hidden in what looks quoted text about figuring out latex later) |
|
A useful notion in Guix is the "build system", which is a package of tools and conventions to manage a build process. Guix has a build system based on autoconf/automake, one based on Python's distutils, etc. Considering that "building" means nothing else than "producing a digital artefact", this can easily be extended to computational science. Running a data analysis is the same as building a data analysis report. Given the current state of the art (which is a mess), I think the best approach would be to allow arbitrary build systems, the condition being that they produce rendered output according to some criteria. Users would be strongly encouraged to use an existing build system rather than make their own, so in the end we'd have a few but not many. Another aspect of Guix build systems worth copying is that the input to a build system is declarative and therefore analyzable. |
|
As food for thoughts. Let's keep simple things simple and hard things possible. Many workflows only require python+cython or R. These cases should not be more complex because of the requirements of other workflows which requires building custom code etc... |
|
Agree with you Antonino. Being able to bring your own docker container to a HPC system, batch queue re: custom software Take a look at On Wed, Feb 24, 2016 at 4:55 PM Antonino Ingargiola <
|
|
Recast project is very workflow oriented. Here's a recent talk focusing on docker, and "parametrized workflows" for the LHC context. Workflow stuff starts around slide 10. @lukasheinrich can do a better job of describing this, but here's a try: We are preparing a document that describes high-level design for executing "parametrized workflows". In the current design there are schedulers that parse the workflow template and the various parameters that are needed to start executing steps in the DAG. @michal-szostakl is working on making this talk to various types of clusters (AWS, carina, google container project, CERN container project, etc.). This produces what we call a "workflow instance" (eg. the specific jobs that ran, their outputs, etc.) and that can be described with something like PROV.
|
|
Hi all, I think the model we came up with can be quite general and in my initial tests it was easy to describe even somewhat complex workflow graphs. The reason we separated the "workflow template" from the "workflow instance" is that this maps better to how usually we think about these workflows. I.e. in our heads we think of a workflow stage as "process all these files from the previous stage in parallel" instead of thinking in terms of very concrete filenames / paths. Also sometimes the full graph is not known ahead of time (which is why we couldn't use snakemake / pydoit / and friends) For the actual workflow instance that @cranmer posted above, this is the graph of the workflow template I intentionally modelled it such that it could be written down somewhat succinctly in a travis-like manner and can be executed locally (as @cranmer mentioned we're working on remote execution as well) |
|
Also i agree with @tritemio. If stuff is really simple, it should stay simple and not be overly complex. If e.g. you can package all your requirements in a single e.g. docker image and run the workflow with you shouldn't need to specify a whole lot more. |
|
this would be the simplest example of a single step process that is parametrizes by an input and output argument. As @ctb said, this more declarative way of specifying the workflow allows for many downstream applications. you can query e.g. what code is used (i.e. what docker images), what the interdependencies of various workflow steps are, what the parameters are etc. (that's what makes it easy for us to visualize) |
|
I like all of the comments here! What about including links to these issues in the proposal? I don't think we want to say we've reached any conclusions yet, and the proposal is due tomorrow, but I think these discussions are incredibly valuable and we can point to them as initial progress. |
|
That is a good idea! 👍 On Sun, Feb 28, 2016 at 4:16 PM C. Titus Brown notifications@github.com
|
|
See also #50 . Note there are two notions of "workflow" being discussed. One is how a user of |
|
Hi, I recently stumbled on http://common-workflow-language.github.io/ and it seems like another workflow specification language, apparently used primarily bio/med fields. Does anyone here have experience with this / know anything about it? Cheers, |
|
Maybe...
https://www.genomeweb.com/informatics/seven-bridges-funds-uc-davis-support-development-standardized-workflow-language
:)
It's kind of a meta specification, and while it's something we should support
I didn't want to bake it into the proposal.
|
|
ugh.. behind a paywall even from NYU network. is there free info on this somewhere? Obviously there is an interest in this across fields in having something like this, which is good. |
|
I’m not premium, so I can’t see that article :-)
|
|
On Mon, Feb 29, 2016 at 10:47:59AM -0800, Lukas wrote:
I can send you a PDF but that doesn't help in general, does it? Anyway, yes, I |
|
That's great. So, I skimmed over that and it seems somewhat similar to our workflow spec. Maybe there is an opportunity there to converge. The one feature, I think that I haven't seen elsewhere, is flexibility in the workflow DAG itself. Our spec allows for extending the graph in certain ways while it is running, which is helpful in cases where the graph structure depends on the outcomes of previous nodes in the graph. |
|
On Mon, Feb 29, 2016 at 10:55:20AM -0800, Lukas wrote:
Sounds like a nice convergence! Drop me an e-mail titus@idyll.org if you |
|
Hello, @mr-c here. I'm the Community Engineer for the #CommonWL. I'm coming up to speed on what you all are doing and I see a lot of crossover. One of my main personal motivations for CWL was that there should be a way to run the complete analysis graph from a paper AND re-mix/re-use it with your own data. Hopefully our 3rd draft of the spec provides much of the functionality needed by that. I'm not sure why @ctb thinks of us as a meta-specification; CWL tool descriptions and workflows made from those tool descriptions are completely runnable on a local machine, in a docker container, or on an academic cluster/grid. We have a chat room if you'd like some real time conversation at https://gitter.im/common-workflow-language/common-workflow-language FYI @betatim we have Docker containers running on HPC systems without root: https://github.com/common-workflow-language/common-workflow-language/wiki/Userspace-Container-Review#getting-userspace-containers-working-on-ancient-rhel |
|
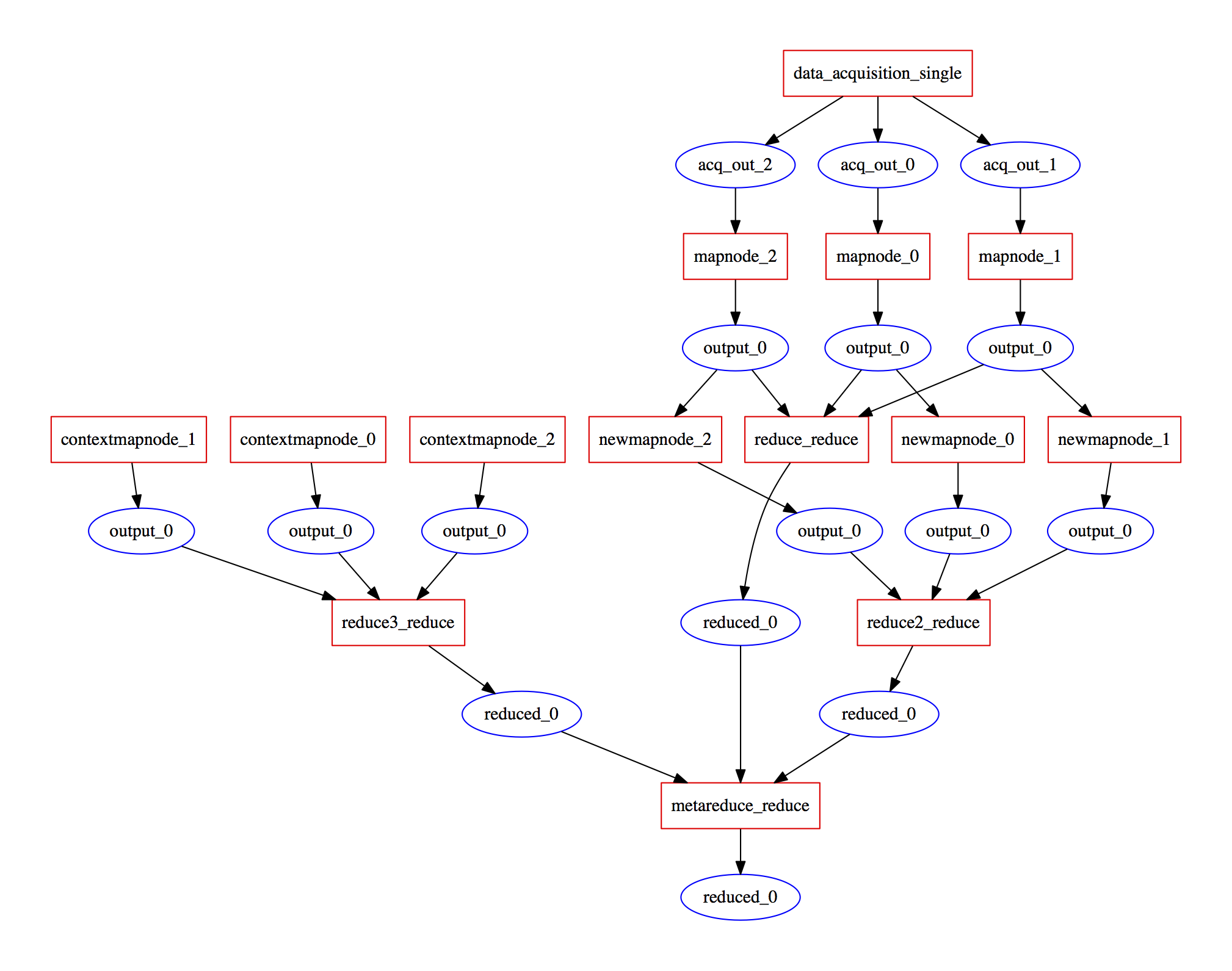
Hi @mr-c, so re-mixing is exactly a point where the flexibility in the graph itself becomes important. Think of this simple type of map-reduce workflow:
now, different input parameters might result in different number of produced files, so the actual graph becomes invocation dependent (though similar structurally between invocations). We solved this in our proposal by allowing for "schedulers" which take a graph -- as executed until this point -- and its invocation parameters to extend the graph with the nodes based on the results up to that point. One approach of course is to hide the parallel computation in a single step that handles all of it, but that goes a bit against the re-usability ideal, since the core component one want to re-use is what happens in each node. Have you encountered these things within the CWL development? |
|
Hello @lukasheinrich Yep, this topic comes up frequently. We currently support the scenario you outlined with our scatter/gather feature: http://common-workflow-language.github.io/draft-3/Workflow.html#WorkflowStep I'm sure that additional dynamic features will be added after our 1.0 release. If we are missing anything, especially derived from the usecases presented in this repo, I really want to hear about it! |
|
yes scatter / gather (which I guess is almost synonymous with map/reduce) is one very common way this graph extensions work, but probably there are more, so we wanted to make this a first-class citizen using the notion of "workflow templates", and "workflow instances". In our JSON-based workflow schema, we allow for arbitrary sub-schemas (which need to be supported by the workflow engine that runs them). This allows custom contributions / workflow patterns to appear organically (maybe curated by a community) Another question: can one run workflows using different docker containers (for each node in the graph) using CWL? If so, how do you describe the environment (which docker container, how to setup a shell environment within the container etc) and how do you coordinate a shared filesystem between those containers? In our case, we allow a list of resources to be listed (such as a network filesystem or a shared host directory), and docker containers can expert to see the work directory at a well-defined path (e.g. |
|
I'm sure we'll add additional dynamic workflow patterns as the standard develops. With CWL you can indeed define a different docker container to use for each tool or step: http://common-workflow-language.github.io/draft-3/CommandLineTool.html#DockerRequirement We are also adding support for giving hints to the local system in the event you'd like to execute a CWL workflow using a traditional HPC cluster. File staging is left as an implementation detail for CWL compliant platforms (some are shared filesystems, many are not). More on the runtime environment: http://common-workflow-language.github.io/draft-3/CommandLineTool.html#Runtime_environment Specific files to be used in computation are specified in the input object, a JSON formatted list of input parameters including file locations. |
|
@lukasheinrich Is there a link for the recast workflow spec? We maintain a (depressingly long) list of other scientific workflow systems at https://github.com/common-workflow-language/common-workflow-language/wiki/Existing-Workflow-systems and I'd like to add y'all. |
|
we're working on a draft right now, hopefully there'll be something presentable soon. |



Outline the envisioned workflow for a scientist. With this we can build a better idea of what needs teaching, blue-printing, etc
First suggestion for a workflow:
openscience initto create a skeletongit init, creates a "sensible"Dockerfileopenscience run <cmd>which executes it inside the docker container.mdwith code blocks that has narrative mixed with steps for reproducing parts of the analysisgit commitall alongopenscience paper(?)(I will edit this entry as we iterate)
The text was updated successfully, but these errors were encountered: